We’ve just taken a major step forward in BioSynapStudio’s core physiology. A new Hodgkin–Huxley (HH) implementation is about to be pushed to GitHub, following a round of testing and validation against canonical models. It’s an important validation milestone in BioSynapStudio’s core physiology. The new Hodgkin–Huxley (HH) implementation has been tested against the canonical equations that have defined computational neuroscience for over seventy years — and the match is exact.
What’s new?
- Pure HH dynamics: The membrane equation has been refactored to follow the original Hodgkin–Huxley formulation directly (
C_m dV/dt = I_inj − ΣI_ion), removing the old threshold state machine. - Ion channel accuracy: Sodium (Na) and potassium (K) channels now use the classic α/β rate equations; the leak channel is truly passive; calcium (Ca) is modelled as high-voltage activated and optional.
- Spike analysis: We’re deprecating the old
PotentialTypein favour ofSpikeStage, with classification handled dynamically bySpikeClassifier. - Synaptic integration: Soma currents now include synaptic/electrode inputs via an overridable
GetExternalCurrent(t). Spikes trigger hillock and STDP events. - Event-driven architecture: Hillock and terminal behaviour now react directly to action peaks, eliminating artificial thresholds.
- Numerical stability: Default timestep is 0.01 ms, with optional sub-stepping and pulse auto-stop to avoid overshoot.
Why it matters
This update brings BioSynapStudio’s spikes into closer alignment with biological reality:
- Spikes peak in the expected range (+30–40 mV) with after-hyperpolarisation below –70 mV.
- Comparisons with Brian2 reference models show sub-millivolt error margins (RMSE).
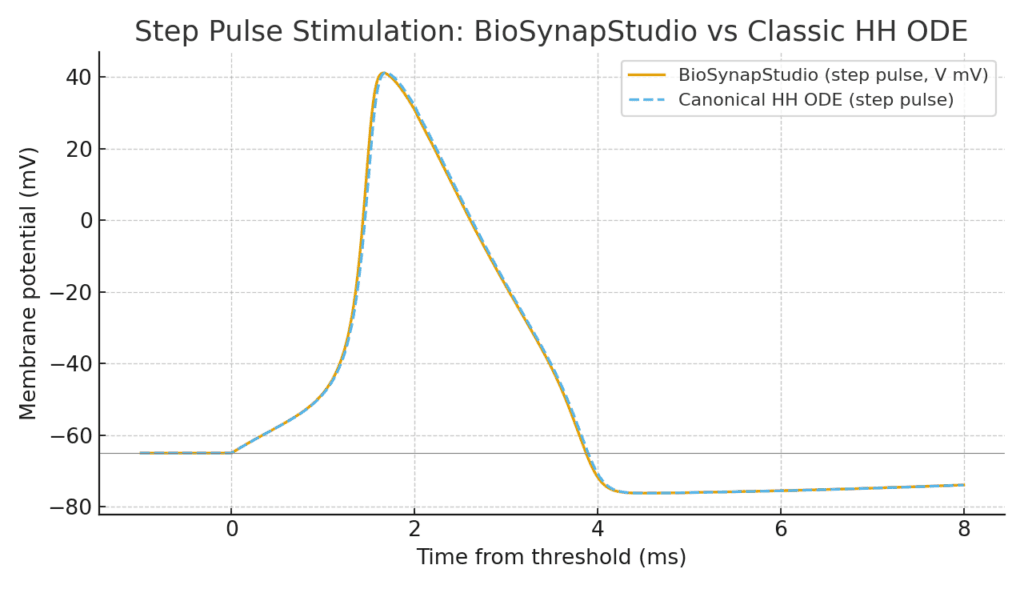
Visual proof
The plot below compares BioSynapStudio’s new HH implementation with the canonical Hodgkin–Huxley model of the squid giant axon.
- The blue dotted line shows the canonical HH ordinary differential equation (ODE) solution, itself derived from the classic squid giant axon data. This has been the gold-standard benchmark in neuroscience for decades.
- The solid orange line is BioSynapStudio’s output under identical conditions.
They are virtually indistinguishable:
This isn’t a heuristic or an approximation. BioSynapStudio is faithfully reproducing the benchmark equations that computational neuroscience is built on. That means:
- Scientific credibility – Researchers can trust that BioSynapStudio spikes are as valid as the reference model they’ve used for decades.
- Platform validation – If we can reproduce HH dynamics with sub-millivolt accuracy, we can build confidently toward more complex cells, synapses, and networks.
- Accessibility – What once required HPC infrastructure can now run on commodity hardware, making biologically faithful models available far more widely.
For scientists, this is the proof that BioSynapStudio isn’t a toy model. For investors and funders, it’s the strongest signal yet that our platform is capable of scaling into multi-cell, network-level, and ultimately full synthetic intelligence applications.
What’s next
We’re considering an optional StageClassifier hook for membrane-only models, and we’ll review whether to adjust default parameters (currently: ENa = +50 mV, EK = –77 mV, EL = –54.387 mV, C_m = 1 µF/cm²).
A short validation report with plots and metrics will be included with the commit. For those partners interested in a deeper dive, early testbed results are available on request.
This milestone confirms what we’ve been working toward: BioSynapStudio isn’t just an idea — it’s already capable of reproducing biology’s most trusted mathematical models, and doing so with precision that stands up to any benchmark.